Photo by Stephen Rossiter
Photo by Stephen Rossiter
Publications
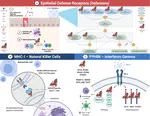
Zoonoses are infectious diseases transmitted from animals to humans. Compared to other mammalian orders, bats are suggested to harbor more zoonotic viruses(Olival et al. 2017). Infections in bats are largely asymptomatic(Schlottau et al. 2020; Guito et al. 2021), suggesting limited tissue-damaging inflammation and immunopathology. To investigate the genomic basis of disease resistance, the Bat1K project generated reference-quality genomes of ten bat species, including potential viral reservoirs. A systematic analysis covering 115 mammalian genomes revealed that signatures of selection in immune genes are more prevalent in bats compared with other mammalian orders. We found an excess of immune gene adaptations in the ancestral chiropteran branch and in many descending bat lineages, highlighting viral entry and detection factors, and regulators of antiviral and inflammatory responses. ISG15, an antiviral gene contributing to hyperinflammation during COVID-19(Perng and Lenschow 2018; Munnur et al. 2021), exhibits key residue changes in rhinolophid and hipposiderid bats. Cellular infection experiments show species-specific antiviral differences and an essential role offor protein conjugation in antiviral function of bat ISG15, separate from its role in secretion and inflammation in humans. Furthermore, in contrast to human ISG15, ISG15 of most rhinolophid and hipposiderid bats has strong anti-SARS-CoV-2 activity. Our work reveals molecular mechanisms contributing to viral tolerance and disease resistance in bats.
Ariadna E. Morales,
Yue Dong,
Thomas Brown,
Kaushal Baid,
Dimitrios - Georgios Kontopoulos,
Victoria Gonzalez,
Zixia Huang,
Alexis-Walid Ahmed,
Leon Hilgers,
Sylke Winkler,
Graham M. Hughes,
Xiaomeng Li,
Bogdan M. Kirilenko,
Paolo Devanna,
Tanya M. Lama,
Yomiran Nissan,
Martin Pippel,
Liliana M. Dávalos,
Sonja C. Vernes,
Sebastien J. Puechmaille,
Stephen J. Rossiter,
Yossi Yovel,
Joseph B. Prescott,
Andreas Kurth,
David A. Ray,
Burton K. Lim,
Eugene Myers,
Emma C. Teeling,
Arinjay Banerjee,
Aaron T. Irving,
Michael Hiller
A de novo assessment of TE content in 248 mammals finds informative trends in mammalian genome evolution.
Austin B. Osmanski,
Nicole S. Paulat,
Jenny Korstian,
Jenna R. Grimshaw,
Michaela Halsey,
Kevin A.M. Sullivan,
Diana D. Moreno-Santillán,
Claudia Crookshanks,
Jacquelyn Roberts,
Carlos Garcia,
Matthew G. Johnson,
Llewellyn D. Densmore,
Richard D. Stevens,
Zoonomia Consortium,
Jeb Rosen,
Jessica M. Storer,
Robert Hubley,
Arian F.A. Smit,
Liliana M. Dávalos,
Kerstin Lindblad-Toh,
Elinor K. Karlsson,
David A. Ray
Horizontal transfer of transposable elements is an important mechanism contributing to genetic diversity and innovation. Bats (order Chiroptera) have repeatedly been shown to experience horizontal transfer of transposable elements at what appears to be a high rate compared to other mammals. We investigated the occurrence of horizontally transferred DNA transposons involving bats. We found over 200 putative horizontally transferred elements within bats; sixteen transposons were shared across distantly related mammalian clades and two other elements were shared with a fish and two lizard species. Our results indicate that bats are a hotspot for horizontal transfer of DNA transposons. These events broadly coincide with the diversification of several bat clades, supporting the hypothesis that DNA transposon invasions have contributed to genetic diversification of bats.
Nicole S. Paulat,
Jessica M. Storer,
Diana D. Moreno-Santillán,
Austin B. Osmanski,
Kevin A.M. Sullivan,
Jenna R Grimshaw,
Jennifer Korstian,
Michaela Halsey,
Carlos Garcia,
Claudia Crookshanks,
Jaquelyn Roberts,
Arian F.A. Smit,
Robert Hubley,
Jeb Rosen,
Emma C. Teeling,
Sonja C. Vernes,
Eugene Myers,
Martin Pippel,
Thomas Brown,
Michael Hiller,
Zoonomia Consortium,
Danny Rojas,
Liliana M. Dávalos,
Kerstin Lindblad-Toh,
Elinor K. Karlsson,
David A. Ray
While evolvability of genes and traits may promote specialization during species diversification, how ecology subsequently restricts such variation remains unclear. Chemosensation requires animals to decipher a complex chemical background to locate fitness-related resources, and thus the underlying genomic architecture and morphology must cope with constant exposure to a changing odorant landscape; detecting adaptation amidst extensive chemosensory diversity is an open challenge. In phyllostomid bats, an ecologically diverse clade that evolved plant-visiting from an insectivorous ancestor, the evolution of novel food detection mechanisms is suggested to be a key innovation, as plant-visiting species rely strongly on olfaction, supplementarily using echolocation. If this is true, exceptional variation in underlying olfactory genes and phenotypes may have preceded dietary diversification. We compared olfactory receptor (OR) genes sequenced from olfactory epithelium transcriptomes and olfactory epithelium surface area of bats with differing diets. Surprisingly, although OR evolution rates were quite variable and generally high, they are largely independent of diet. Olfactory epithelial surface area, however, is relatively larger in plant-visiting bats and there is an inverse relationship between OR evolution rates and surface area. Relatively larger surface areas suggest greater reliance on olfactory detection and stronger constraint on maintaining an already diverse OR repertoire. Instead of the typical case in which specialization and elaboration are coupled with rapid diversification of associated genes, here the relevant genes are already evolving so quickly that increased reliance on smell has led to stabilizing selection, presumably to maintain the ability to consistently discriminate among specific odorants — a potential ecological constraint on sensory evolution.
Laurel R. Yohe,
Matteo Fabbri,
Daniela Lee,
Kalina T.J. Davies,
Thomas P. Yohe,
Miluska K.R. Sanchez,
Edgardo M. Rengifo,
Ronald P. Hall,
Gregory L. Mutumi,
Brandon P. Hedrick,
Alexa Sadier,
Nancy B. Simmons,
Karen E. Sears,
Elizabeth R. Dumont,
Stephen J. Rossiter,
Bhart-Anjan Bullar,
Liliana M. Dávalos
Comprising more than 1,400 species, bats possess adaptations unique among mammals including powered flight, unexpected longevity, and …
Diana D. Moreno-Santillán,
Tanya M. Lama,
Yocelyn Gutiérrez Guerrero,
Alexis M. Brown,
Paul Donat,
Huabin Zhao,
Stephen Rossiter,
Laurel R. Yohe,
Joshua Potter,
Emma C. Teeling,
Sonja Vernes,
Kalina T.J. Davies,
Eugene Myers,
Graham M. Hughes,
Zixia Huang,
Federico G. Hoffmann,
Angelique P. Corthals,
David Ray,
Liliana M. Dávalos
Bats possess extraordinary adaptations, including flight, echolocation, extreme longevity and unique immunity. High-quality genomes are crucial for understanding the molecular basis and evolution of these traits. Here we incorporated long-read sequencing and state-of-the-art scaffolding protocols1 to generate, to our knowledge, the first reference-quality genomes of six bat species (Rhinolophus ferrumequinum, Rousettus aegyptiacus, Phyllostomus discolor, Myotis myotis, Pipistrellus kuhlii and Molossus molossus). We integrated gene projections from our ‘Tool to infer Orthologs from Genome Alignments’ (TOGA) software with de novo and homology gene predictions as well as short- and long-read transcriptomics to generate highly complete gene annotations. To resolve the phylogenetic position of bats within Laurasiatheria, we applied several phylogenetic methods to comprehensive sets of orthologous protein-coding and noncoding regions of the genome, and identified a basal origin for bats within Scrotifera. Our genome-wide screens revealed positive selection on hearing-related genes in the ancestral branch of bats, which is indicative of laryngeal echolocation being an ancestral trait in this clade. We found selection and loss of immunity-related genes (including pro-inflammatory NF-κB regulators) and expansions of anti-viral APOBEC3 genes, which highlights molecular mechanisms that may contribute to the exceptional immunity of bats. Genomic integrations of diverse viruses provide a genomic record of historical tolerance to viral infection in bats. Finally, we found and experimentally validated bat-specific variation in microRNAs, which may regulate bat-specific gene-expression programs. Our reference-quality bat genomes provide the resources required to uncover and validate the genomic basis of adaptations of bats, and stimulate new avenues of research that are directly relevant to human health and disease1.
David Jebb,
Zixia Huang,
Martin Pippel,
Graham M. Hughes,
Ksenia Lavrichenko,
Paolo Devanna,
Sylke Winkler,
Lars S. Jermiin,
Emilia C. Skirmuntt,
Aris Katzourakis,
Lucy Burkitt-Gray,
David A. Ray,
Kevin A. M. Sullivan,
Juliana G. Roscito,
Bogdan M. Kirilenko,
Liliana M. Dávalos,
Angelique P. Corthals,
Megan L. Power,
Gareth Jones,
Roger D. Ransome,
Dina Dechmann,
Andrea G. Locatelli,
Sebastien J. Puechmaille,
Olivier Fedrigo,
Erich D. Jarvis,
Mark S. Springer,
Michael Hiller,
Sonja C. Vernes,
Eugene W. Myers,
Emma C. Teeling